ORMIR-MIDS as a Standard Specification#
The ORMIR-MIDS specifications are a simple and intuitive way to organize MSK imaging data. They are an extension of the specifications of the Brain Imaging Data Structure (BIDS) and the Medical Imaging Data Structure (MIDS)
In this page, you will find:
1. Structure and naming of folders and files
2. Supported imaging modalities
3. Current and future work
Structure and naming of folders and files#
Folder structure#
The folder structure is hierarchical and composed of 4 levels of nested folders:
Dataset → Subject → Session → Imaging Method
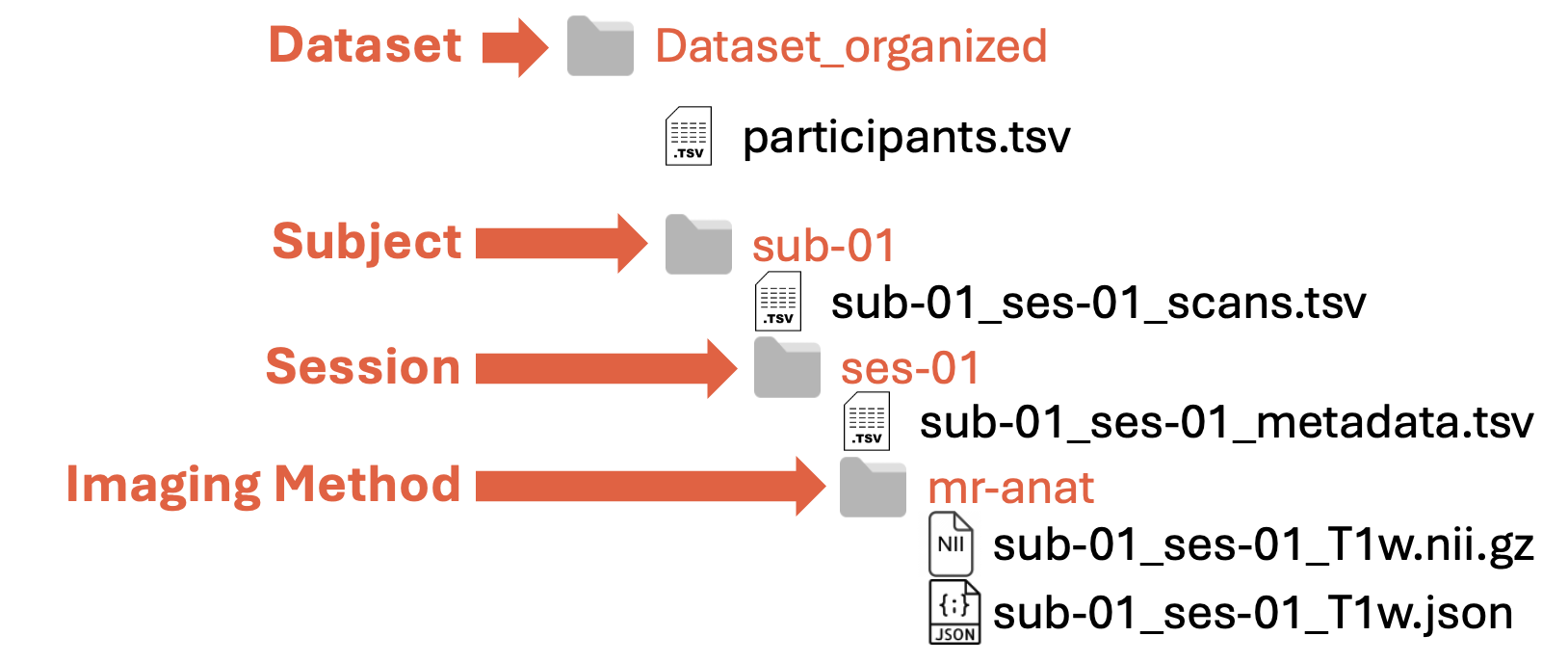
Dataset: root (or parent) folder containing all the MSK data (E.g.,
Dataset_organized)Subject: folder(s) containing all the data about a single subject (E.g.,
sub-01for the first subject)Session: folder(s) containing the data of an acquisition session at a specific time point(E.g.,
ses-01for baseline acquisitions)Imaging Method: folder(s) containing the data of a specific acquisition. (E.g.,
mr-anatfor anatomical MR images). You can find the currently supported imaging methods in this table
Filesystem structure#
Within each folder or subfolder, there are specific files:
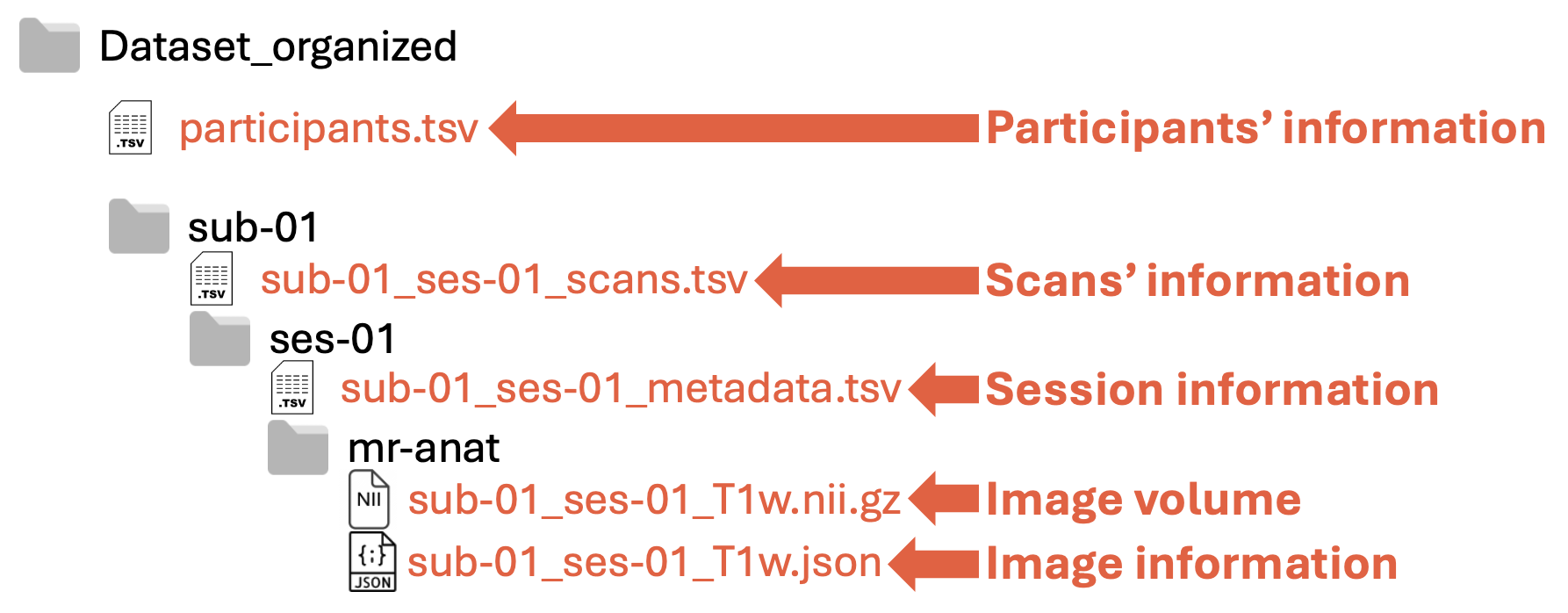
Participants’ information: definition under construction!
Scans’ information: definition under construction!
Session information: definition under construction!
Image volume: 3D image
Image information: Header of the dicom images in
.jsonformat
Folder naming#
Folders have precise names:
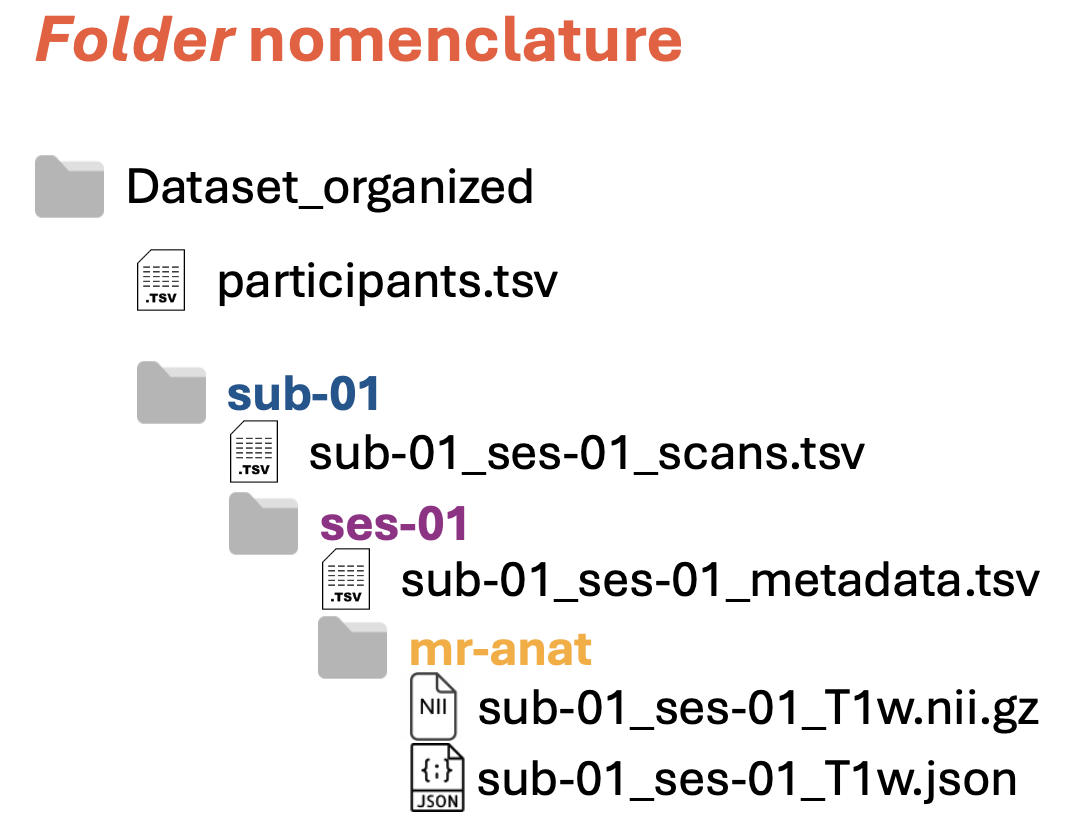
Folder names referring to subjects and sessions are composed of two parts:
suborses, indicating subject (blue in the figure above) or session (purple in the figure above)<ID>of the subject or of the session (e.g.,01)
Note: The two parts are separated by dash (
-)Folder names referring to image modalities (yellow in the figure above) can have various names, such as
mr-anat,ct, etc. For a list of the available names, see the second column (called Folder name) in the table below
File naming#
Files have precise names:
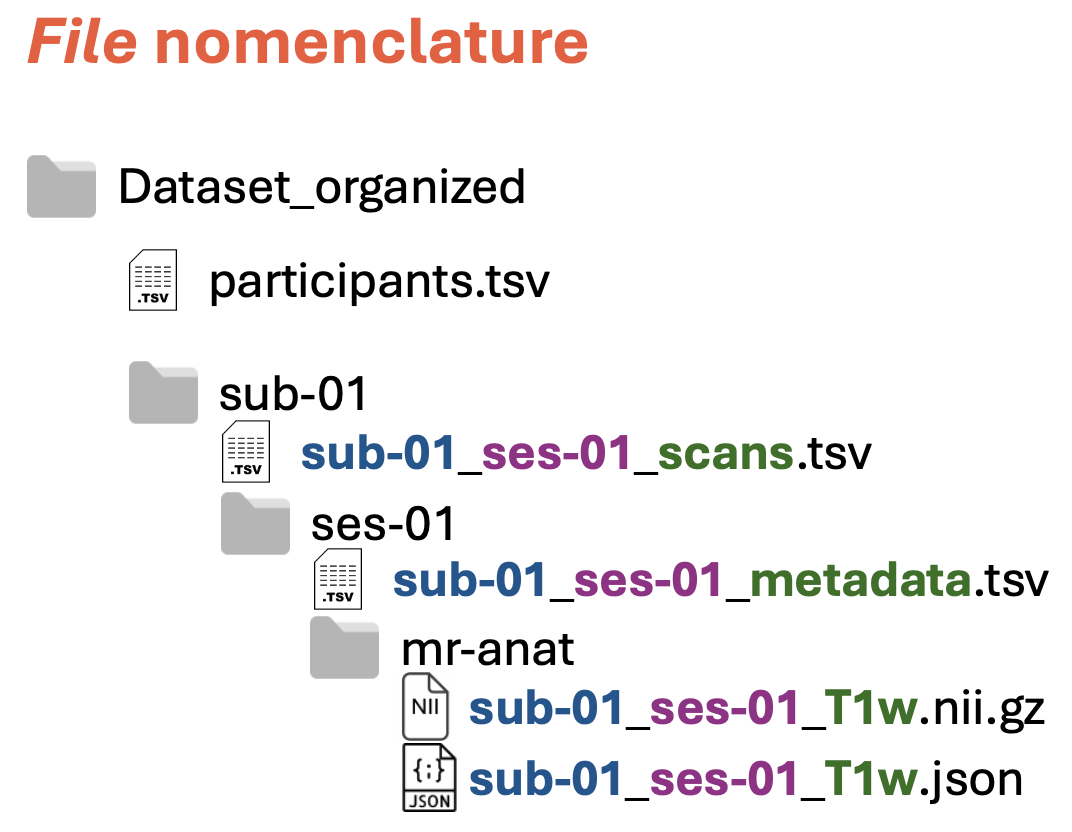
Each file name is composed of three parts, each of them containing information about:
Subject, such as
sub-01(blue in the figure above)Session, such as
ses-01(purple in the figure above)File content, such as
scans,metadata, etc. (green in the figure above). For the images, you can find the currently supported file types in the table below
Note: The three parts are separated by underscore (
_)
General JSON Fields#
The JSON fields in the following table are common to all acquisition types of a specific modality. Refer to the section below for specific acquisition types within a modality. The table columns specify:
Imaging method: the image acquisition method, such as
computed tomography,MR imaging, etc.Folder names: name of the output folder containing the images (e.g.,
mr-anat,ct, etc.). See the folder naming specifications for more detailsFile name suffix: suffix of the file name of the image volume (e.g.,
mese,megre, etc.). See the file naming specifications for more detailsImage volume: dimension (
2D,3D, or4D) and directions (x,y,z,echo) of the obtained image volume (i.e.,nii.gzfiles)JSON fields: information that the dicom header must have to be able to transform the data into the ORMIR-MIDS structure. Fields in bold are mandatory. Units (if available) are in square brackets.
Imaging method |
Folder name(s) |
File name suffix |
Image volume |
JSON fields |
|---|---|---|---|---|
Computed tomography |
ct |
N/A |
3D (x,y,z) |
XRayEnergy [kVp]
|
Computed / plain radiography |
cr |
N/A |
2D (x,y) |
ExposureTime [ms]
|
MRI |
mr-anat/mr-quant |
[various] |
3D, 4D, or 5D |
EchoTime [ms] |
MRI: T1w / T1w-FS / T2w / T2w-FS |
mr-anat |
t1w / t1w-fs / t2w / t2w-fs |
3D (x,y,z) |
|
MRI: Quantitative T1 / T2 / wT2 |
mr-quant |
t1 / t2 / wt2 |
3D (x,y,z) |
Acquisition-specific specifications#
Current and future work#
We are working on converters for:
More imaging modalities, such as HR-pQCT, MRI Diffusion
Derived data, such as segmentation labels
Here are the specs of some imaging modalities we will support:
Diffusion
NIfTI structure: 4D (x,y,z,direction)
Filename suffix: diff
Folder: diff ??
JSON required fields:
MixingTime in ms
EncodingDirection (array of 3D vectors). The norm of the vector is the b-value. The normalized vector indicates the direction of the diffusion gradient in patient coordinates.
Segmentation labels
NIfTI structure:
3D (x,y,z) with integer values, from 0 to N, each value corresponding to a different label
4D (x,y,z,label) stack of binary (0/1) values, each dimension corresponding to a label (one-hot encoding)
Filename suffix: seg ??
Folder: seg ??
JSON required fields:
Labels (array of strings). List of the labels represented in the masks. The first value in the list corresponds to either a gray level of 0 or to the 1st volume in the fourth dimension. E.g.
["Background", "SOL", "VM", "VL"]Note: the string representation of the labels must follow a standardized format. While it is possible that the same anatomical structure is represented by different labels (e.g.
SOLorSoleus), the labels must be known. This allows flexibility in the implementation of segmentation tools, while keeping easy interoperability because all values are easily convertible. A list of standardized labels is visible here. Please contact Francesco Santini if you would like to add your own definitions.
